Synthetic sex in yeast promises safer medicines for people
By exploiting the way yeast cells mate, researchers have figured out a quicker, easier way to identify on- and off-target drug interactions.

Our old friend Saccharomyces cerevisiae – the yeast that’s helped people bake bread and brew beer for millennia – has just had its sex life upgraded.
Bioengineers at the University of Washington have reprogrammed the mating habits of this single-celled organism, letting the fungus get it on like never before. The result? A sexual revolution that could lead scientists to safer future medicines.
Yeast as lab guinea pig
We already rely on yeast for a lot more than just fermented food. Much of our modern understanding of genetics and cell biology has come from careful study and manipulation of the fungus.
Scientists and drug designers love working with yeast because of its rapid cell cycle (a new generation is born every 90 minutes) and the relative ease with which its genes can be tweaked. Even human genes and genes encoding protein-based drugs can be spliced in, allowing researchers to study them in the lab in detail. Anti-cancer drugs have been optimized this way.
One of the most popular techniques for this type of biomolecular research is known as yeast surface display. Using this method, a gene can be added to yeast and the protein that results will appear on the easily accessible outer surface of the cell. With a new protein displayed on the surface, researchers can easily determine what other biomolecules the protein interacts with.
This method, pioneered in the laboratory of Dave Wittrup, exploits aspects of the fungus’ sexual machinery.
Yes, even single-cell microbes can have sex. But as is often the case outside the animal kingdom, the way DNA gets swapped can seem unusual to human observers.
Fungal fornication
The terms “male” and “female” don’t really apply to budding yeast. Instead of forming sperm or eggs, the sex cells of yeast all look the same – like tiny, single-cell blobs. What makes two yeast blobs able to sexually reproduce are their so-called mating types.
The proteins that decorate the outside of a yeast sex cell, or gamete, determine that cell’s mating type. Put on copies of one protein and you’re one mating type; swap them out for a different protein and you’re the other. Agglutination (the unsexy term for yeast sex) only happens when the surface proteins of yeast gametes from opposite mating types interact.

Inspired by this molecular precision, a team of synthetic biologists led by University of Washington graduate student David Younger realized they could convert the natural yeast mating system into a new tool that would let them precisely measure molecular interactions at a much larger scale.
Though tiny and difficult to measure, molecular interactions are a big deal in drug design. Virtually every drug works via specific interactions with its target, and drugs that bind where they shouldn’t can be lethal.
Some experts blame off-target interactions for last year’s failed phase III clinical trial of Alnylam Pharmaceuticals’ revusiran, an RNA drug designed to treat a rare heart disease. Nineteen people died before the trial was called off, and the company’s stock took a US$3 billion hit.
Figuring out whether a new drug binds what it’s supposed to is relatively easy; figuring out whether it binds anything else in our cells is tough. Established laboratory techniques like yeast surface display have helped scientists screen new drugs for potentially dangerous off-target interactions before they make it to clinical trials, but that technique lets you look for off-target interactions only one at a time. Younger’s team envisioned a way to test hundreds of drugs against thousands of potential targets, all by redesigning yeast sex.
Redesigning yeast sex with multiple mating types
To start, Younger needed a way to precisely measure mating efficiency in lab-grown yeast. Perfect efficiency would mean every cell that could fuse would do so. The more efficient the mating, the better matched the two mating types.
The team linked genetically encoded fluorescent markers – one blue, one red – to each of the natural yeast mating types. That made it simple to measure mating efficiency for a whole yeast population: They could just count the cells that stayed blue or red (unmated) versus those that turned purple (mated). It turns out for typical yeast grown in the lab, mating efficiency is around 60 percent.
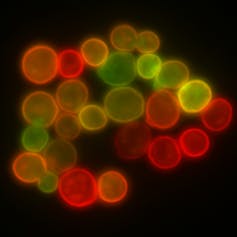
The team then deleted the natural agglutination proteins and replaced them with a pair of foreign proteins known to interact weakly. The mating efficiency dropped tenfold to 5.7 percent. They swapped in another pair and saw it rise to 19 percent. When they tried a third pair of proteins known to interact with much higher affinity, mating efficiency rose to 51.6 percent – close to what was seen in natural agglutination.
Just by tracking mating efficiency, the team could tell how strongly any two protein molecules interact. When they checked a pair of proteins that shouldn’t interact at all, mating efficiency was a meager 0.2 percent.
Now, instead of just the two natural mating types, scientists can quickly engineer thousands of “sexes” by coaxing individual yeast to decorate the outside of their cells with new, human-specified proteins. If a pair of new mating types are sexually compatible – meaning the proteins decorating their cell surfaces interact – their offspring will rise in number. By tallying up each genetically distinct offspring in a tube not much bigger than a thimble, thousands of potential molecular interactions can be quantified.
Improving drug safety
To show that their new tool could aid in drug development, the team generated 1,400 distinct variants of an emerging anti-cancer drug known as XCDP07. The drug is supposed to disrupt the unrestrained growth of cancer cells by binding specific cellular targets, but as with every drug, significant off-target interactions could render it useless. By mixing yeast displaying different versions of the drug with other yeast displaying human proteins, the team was able to identify versions of XCDP07 which only bound to the intended target.
Younger is working to get his new tool into the hands of more scientists. He’s already gifted his engineered yeast strains to eager researchers at Stanford, Yale, UCSD and beyond. Concerns over the cost and safety of emerging drugs have motivated him to start a company – funded by scientific grants, not investors – to turn his results into the next generation of medicines. Younger says the goal is to provide “comprehensive preclinical drug screening, rather than the current practice of screening a very small subset of possible off-target interactions.”
The next blockbuster drugs may owe a debt to yeast and their mating practices. Who says you can’t teach an old fungus new tricks?
Ian Haydon is a researcher at the Institute for Protein Design at the University of Washington.
Read These Next
How Homeland Security’s subpoenas and databases of protesters threaten the ‘uninhibited, robust, and
It’s difficult to measure what is lost when an opinion is never voiced and impossible to catalogue…
Menstrual pads and tampons can contain toxic substances – here’s what to know about this emerging he
Heavy metals, phthalates and other potentially harmful chemicals have been detected in a range of menstrual…
Last nuclear weapons limits expired – pushing world toward new arms race
The expiration of the New START treaty has the US and Russia poised to increase the number of their…






