Gene-editing technique CRISPR identifies dangerous breast cancer mutations
Mutations in BRCA genes are linked to the early onset of breast and ovarian cancers. But the effect of most mutations is unclear. Now new research can distinguish harmless from dangerous mutations.
More than 1 million women have had genetic testing of BRCA1 and BRCA2, genes in which mutations can dramatically increase the risk for early onset breast and ovarian cancer. But for many women the test results have been ambiguous. That’s because it’s not clear where certain genetic variations are harmless or cause cancer.
BRCA1 was amongst the first cancer predisposition genes discovered, and it has been studied for over 20 years. The gene produces a protein that repairs DNA damage, which might otherwise lead to the formation of tumors. Since its discovery, researchers and clinicians have identified many genetic variations in BRCA1, but for most of these, we are unable to tell whether they impair function of the gene – raising the risk of cancer – or whether they are perfectly harmless.
Our research team works in the emerging field of genomic medicine, which uses an individual’s genetic information to prescribe care. We recognized that such “variants of uncertain significance” limited the utility of genetic testing and the prospects for genomic medicine. We know that problem is likely to get worse, as the number of uncertain variants in BRCA1 and other “medically actionable” genes is expected to grow exponentially as genetic testing is expanded to entire populations.
In a study, we set out to apply CRISPR genome editing to solve the challenge posed by these variants of uncertain significance. CRISPR has tremendous potential because the technology allows researchers like us to tinker with human genes. CRISPR allows us to make very specific changes, “edits” to our DNA – thus the phrase, “genome editing.”
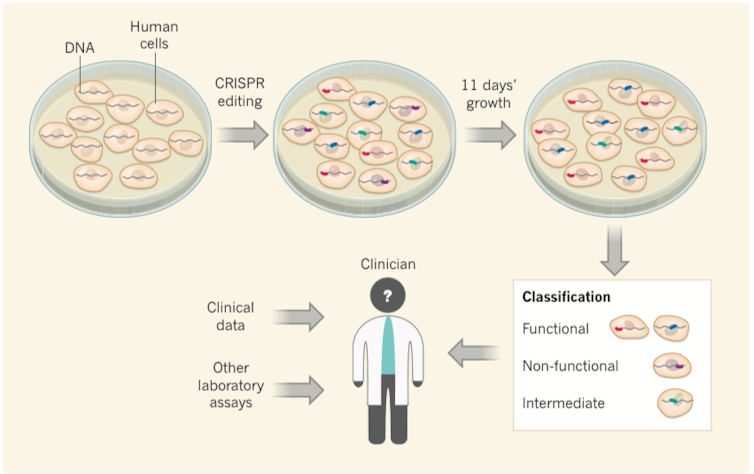
Although there are many studies that are attempting to use CRISPR to treat disease, it can also be used to introduce specific mutations into human cells that grow in a dish, for the purposes of studying what effects these mutations have on the cell – for instance, whether or not they cause a gene to malfunction.
In our study, we used CRISPR genome editing to deliberately engineer some 4,000 different variants of the BRCA1 gene in human cells, nearly all possible variants in the most important regions of this gene. Importantly, the survival of the human cells that we used is dependent on intact function of the BRCA1 gene. As a consequence, the cells containing mutations that disrupted the function of the BRCA1 gene were unable to survive. On the other hand, the cell containing mutations that had no effect on the function of the BRCA1 gene were just fine. Using DNA sequencing, we tracked which mutations were associated with cell death versus cell survival.
When we compared the mutations that caused cell death to variants that are known to increase cancer risk, we noticed that they were the same. This gave us the confidence to say that the behavior of these variants in the cells in the dish was predictive of cancer risk in humans.
Although scientists have used laboratory assays to test variants in BRCA1 for many years, our work is different for three reasons.
First, we tested many more variants than have ever been tested, including thousands that have never been observed before but almost certainly exist in at least hundreds of living humans.
Second, historically BRCA1 variants have been tested in genes taken “out of context” – specifically, studying only the DNA sequences that encode the BRCA1 protein, rather than the surrounding sequences that regulate how it is expressed. CRISPR allows us, for the first time, to create and test the mutations in the human genome itself.
Finally, for the hundreds of BRCA1 variants seen in patients where we do have a good sense of whether or not they increase risk of breast and ovarian cancer, our predictions based on our CRISPR studies are nearly perfectly accurate. That is, the variants compatible with cell survival in our assay are benign in patients, while the variants that impair cell survival in our assay cause cancer risk. This gives us confidence in our predictions for other variants that have never before been observed but inevitably will be, particularly as more and more women are screened for mutations in this gene.
Because of this strong agreement with “gold standard” data derived from human studies, we predict our results can be used to provide better answers to women with challenging-to-interpret variants in BRCA1. This includes many women that have an elevated risk of cancer, but would previously have been missed by genetic testing. To these women, this knowledge of what their mutations mean may critically inform the medical care that they receive.
Jay Shendure receives funding from Brotman Baty Institute for Precision Medicine, National Institutes of Health, Allen Frontiers Foundation, Howard Hughes Medical Group. The authors are co-inventors on a patent application related to the saturation genome editing method, and the scores described here require a license from the University of Washington for for-profit or commercial use.
Lea Starita is employed by the Brotman Baty Institute for Precision Medicine.
Greg Findlay does not work for, consult, own shares in or receive funding from any company or organization that would benefit from this article, and has disclosed no relevant affiliations beyond their academic appointment.
Read These Next
Artists and writers are often hesitant to disclose they’ve collaborated with AI – and those fears ma
Whether they’re famous composers or first-year art students, creators experience reputational costs…
1 protein to rule them all – why crowning the protein that makes jellyfish glow green as a model can
Researchers have been studying tens of thousands of proteins and even more variations without a yardstick…
Baptists have helped shape debate about religious freedom for over 400 years – up to today’s 10 Comm
The phrase ‘separation of church and state’ dates back to a letter from Thomas Jefferson to a Baptist…