3D scans of bat skulls help natural history museums open up dark corners of their collections
Museums' collections are a priceless resource for scientists, but they're not easy to access. Digitizing specimens – like the 700 bat skulls the author studied – is a way to let everyone in.
Picture a natural history museum. What comes to mind? Childhood memories of dinosaur skeletons and dioramas? Or maybe you still visit to see planetarium shows or an IMAX feature? You may be surprised to hear that behind these public-facing exhibits lies a priceless treasure trove that most visitors will never see: a museum’s collections.
Far from being forgotten, dusty tombs, as is sometimes the perception, these collections host the very cutting edge of research on life on this planet. The sheer scale of some of the largest collections can be staggering. The Smithsonian National Museum of Natural History, for instance, houses over 150 million specimens. Even a smaller academic institution, like the Research Museums Center of the University of Michigan, houses a labyrinth of specimen vaults, preserving millions of skeletons, fossils, dried plant material and jarred organisms.
Most importantly, poring over this wealth of knowledge at any given time are active researchers, working to unravel the intricacies of Earth’s biodiversity. At the University of Michigan, where I received my Ph.D. in ecology and evolutionary biology, I worked nestled among these skeletons, fossils and other natural treasures. These specimens were critical to my research, as primary records for the natural history of the world.
Yet despite the incalculable value of these collections, I often wondered about how to make them more accessible. A project to digitally scan hundreds of bat skulls was one way to bring specimens that would look at home in an antique Victorian collection straight to the forefront of 21st-century museum practices.

A valuable resource, largely hidden from view
By researching variation among and within collection specimens, biologists have uncovered many ecological and evolutionary mysteries of the natural world. For instance, a recent study on bird specimens traced the increasing concentration of atmospheric black carbon and its role in climate change over more than a century. Scientists can collect ancient DNA from specimens and gather information about historical population levels and healthy genetic diversity for organisms that are now threatened and endangered.
My own research on global bat diversity used hundreds of museum specimens to conclude that tropical bats coexist more readily than many biologists expect. This finding fits with an overall pattern across much of the tree of life where tropical species outnumber their temperate cousins. It may also help explain why in many parts of Central and South America, bats are among the most abundant and diverse mammals, period.
However, research on these specimens often requires direct access, which can come at a steep price. Researchers must either travel to museums, or museums must ship their specimens en masse to researchers - both logistical and financial challenges. Museums are understandably wary of shipping many specimens that are truly irreplaceable - the last evidence that some organisms ever existed in our world. A museum’s budget and carbon footprint can quickly balloon with loans. And as physical specimens cannot be in more than one location at once, researchers may have to wait an indefinite amount of time while their materials are loaned to someone else.
CT scanning bat skulls
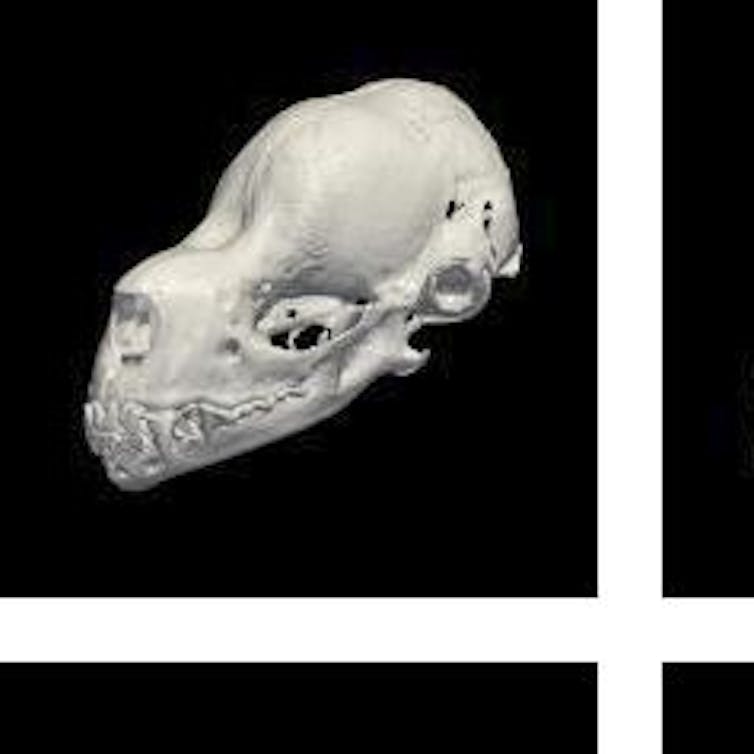
I have tried to tackle these issues of access with my collaborators Daniel Rabosky and Erin Westeen using micro-CT technology. Just like with medical CT scanning, micro-CT uses X-rays to digitize objects without damaging them – in our case, these scans occur at the fine scale of millionths of meters (micrometers). This means micro-CT scans are incredibly accurate at high resolutions. Even very tiny specimens and parts are preserved in vivid detail.
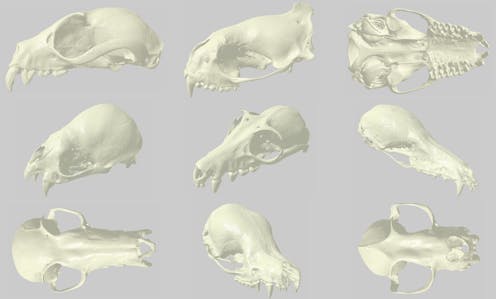
For my Ph.D. research, we used micro-CT scanning to digitize nearly 700 individual bat skulls from our museum’s collection. With estimates of about 1,300 described species, bats represent about 25 to 30 percent of modern mammal species, second only to rodents. However, one of the reasons researchers have long been fascinated by bats is their immense diversity of behavior and function in nature. Much of this ecological diversity is encoded in their skulls, which vary broadly in shape and size.
At the Michigan School of Dentistry’s micro-CT facility, we scanned every bat skull at high resolutions. Each scan produced hundreds of thousands of images per specimen - each image a tiny cross-section of an original skull. With these “stacks” of cross-sections, we then reconstructed 3D surfaces and volumes. In essence, we recreated a 3D “digital specimen” from each of the roughly 700 originals.
Digital specimens open doors
In partnership with MorphoSource at Duke University, we’ve since published our digital specimens within an open-access repository for researchers, educators and students. Each digital specimen is associated with the same identifying data as its original, enabling research without travel or shipment. Even better, many delicate parts can be digitally dissected without fear of irreparable damage. Digital specimens can even be 3D-printed at varying scales for use in educational settings and museum exhibits.
My colleagues Dan and Erin have continued to expand these efforts to other vertebrates at our museum. Our hope is that the broader scientific community will embrace open-access digital specimen data in much the same way that digital, publicly available genetic data has been adopted across biology. Digitization can expand the reach of each museum, especially as scanning prices drop and open-access micro-CT software becomes more practical.
This digital revolution comes at time when many natural history museums are endangered. Around the globe, museums are hamstrung by budget cuts and decades of neglect, with devastating consequences.
One way to revitalize museums is to embrace digital missions that preserve priceless data and promote global collaboration. Far from making physical collections obsolete, digitization can modernize natural history museums, as it has with libraries and other museums of art, history and culture. The originals will always be there for those looking to dive deep into natural history. The digital wing can instead invite curiosity and questions from sources most museums could never dream of otherwise reaching.
In my earliest days as a biologist, I was plagued by common researcher worries. What was going to happen to all of my data? Who else would ever see it? Scientists never know what new life may be breathed into our basic research after years, decades, centuries. I think about the hundreds of past scientists who unknowingly contributed data to my own research, spanning nearly 130 years and six continents of expeditions.
By digitizing their earlier efforts, my colleagues and I ensured that they can reach broad audiences, far beyond what they likely imagined. No longer should the potential impact of any specimen be restricted by the walls and constraints of any one museum. Instead, museums can throw their doors open to a digital future, inviting anyone into the endless wonders of the natural world.
Jeff J. Shi received funding for this research from the National Science Foundation's Division of Environmental Biology, in the form of a Doctoral Dissertation Improvement Grant co-awarded to Dr. Daniel L. Rabosky.
Read These Next
Supreme Court to consider whether local governments can make it a crime to sleep outside if no insid
Legal precedents hold that criminalizing someone for their status, such as being homeless, is cruel…
More climate-warming methane leaks into the atmosphere than ever gets reported – here’s how satellit
Methane is a potent greenhouse gas that can leak from oil and gas wells, pipelines and landfills. Satellites…
Grizzly bear conservation is as much about human relationships as it is the animals
Whether people are hunters can have a big effect.



